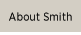Professor of Biological Sciences Stylianos Scordilis compares two, two-dimensional protein gels with Dr. Kalina Dimova ’04, the technical director of the Center for Proteomics, and Ellen Margolis, a biochemistry major who is carrying out her proteomics specials studies project on skeletal muscle cells in culture.
/ Published November 26, 2010
When Army scientists in Maryland and entomologists in Montana recently reported a major breakthrough in identifying the viral and fungal infections that may be killing entire honeybee colonies, the director of Smith’s Center for Proteomics took note. Indeed, it was the science of the protein universe—proteomics—that gave scientists the tools to uncover a new DNA-based virus and establish a connection to the fungus that was afflicting the bee colonies.
According to Stylianos P. Scordilis, professor of biological sciences and the center’s director, one of the fundamental challenges in understanding the molecular basis of life in the 21st century in the post-genomics era, lies in proteomics. Proteomics studies the nature and interactions of proteins in the context of their complex cellular and extracellular environments. “This requires a host of new technologies and techniques.”
Proteomics is a broad set of disciplines aimed at studying and monitoring proteins. This includes work correlating structure with protein function, development of protein separation and protein profiling techniques, and investigation of protein-protein interactions. Proteomics promises to revolutionize medicine and health care by determining the interactions of the hundreds of thousands of proteins found in humans.


Comparison of female muscle protein spots relative to the identical spots in male muscle (green circles o). Spots that are increased in females are circled red and those that are decreased are circled blue.
Scordilis says, “Similarly to the report in The New York Times on proteomics coming to the rescue of honeybees in determining that a viral interaction with a fungus and subsequent infection of the bees caused the problem, proteomics at Smith is answering many questions.”
For example, undergraduates in Professor Scordilis’ laboratory are using these new tools to unravel the gender-specific differences in skeletal muscle. He explains: “Mouse skeletal muscle is extracted in a solution that solubilizes all of the proteins in the muscle. This extract is then separated into a two-dimensional map of the proteins by use of electrophoretic techniques; one separates the proteins based on their native charges (the horizontal dimension) and another separates on the basis of the protein’s molecular weight (the vertical dimension). These two-dimensional gels are then visualized using high-resolution digital imaging; about 800 protein spots from mouse biceps muscle are detected, which make up the muscle proteome map. The spot intensities are measures of the change in the protein amounts. The identities of the protein spots that change more than twofold (up or down), and are statistically significant, are determined by amino acid sequence analysis using liquid chromatography-coupled mass spectrometry.”
“These results show that females demonstrate an abundance decrease in sugar energy metabolism enzymes and in creatine kinase enzyme isoforms, a different energy supply system in muscles. This gender dimorphism in the murine biceps brachii muscle wasn’t known before our work.”
What this points to, notes Scordilis, is the importance of understanding the gender differences of skeletal muscle at the protein (molecular) level in exercise studies.